Examples
Plain AnnData with ScanPy
from metaspace_converter import metaspace_to_anndata
import scanpy as sc
# Download data and convert to an AnnData object
adata = metaspace_to_anndata(
dataset_id="2022-08-05_17h28m56s",
fdr=0.1,
database=("BraChemDB", "2018-01"),
)
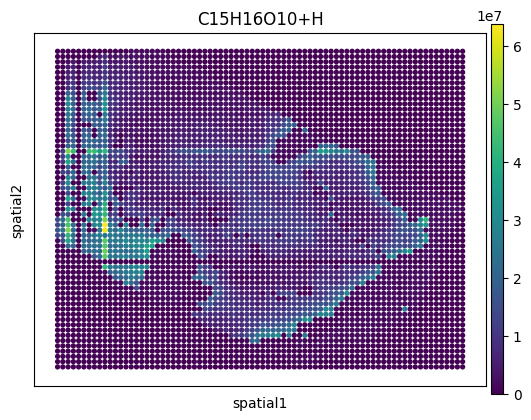
# Visualization with ScanPy
sc.pl.spatial(
adata,
# Choose first ion for visualization
color=adata.var.index[0],
img_key=None,
spot_size=1,
)
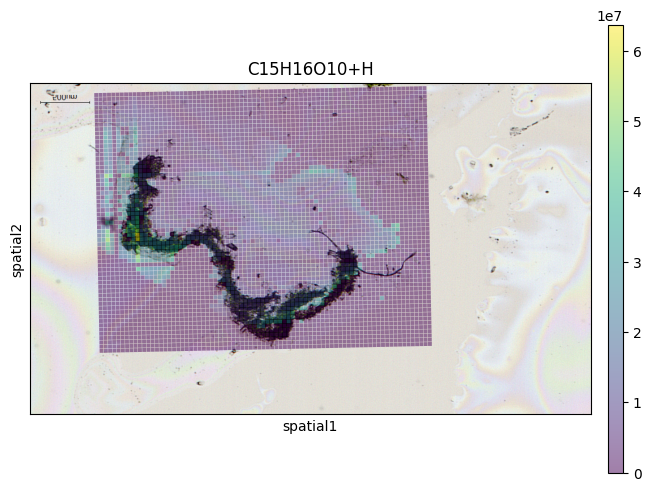
SquidPy
from metaspace_converter import metaspace_to_anndata
import squidpy as sq
# Download dataset with optical background image
adata = metaspace_to_anndata(
dataset_id="2022-08-05_17h28m56s",
fdr=0.1,
database=("BraChemDB", "2018-01"),
add_optical_image=True,
)
sq.pl.spatial_scatter(
adata, color=adata.var.index[0], shape="square", img=True, size=15, alpha=0.5
)
Convert AnnData objects to ion image arrays
If you want to work with the ion images as numpy arrays, the function anndata_to_image_array can convert previously downloaded AnnData objects to numpy arrays.
from metaspace_converter import metaspace_to_anndata, anndata_to_image_array
# Download data
adata2 = metaspace_to_anndata(dataset_id="2023-11-14_21h58m39s", fdr=0.1)
ion_images = anndata_to_image_array(adata2)
# 20 ion images of shape 130x143
print(ion_images.shape)
(20, 130, 143)
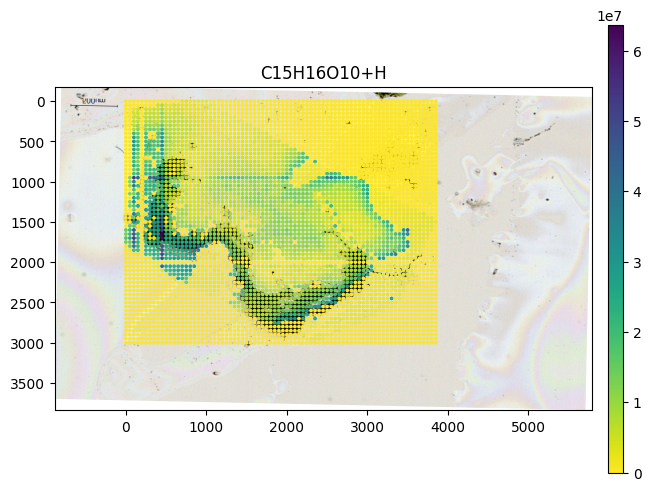
SpatialData
Here using a reversed colormap which better represents intense values on bright background.
from metaspace_converter import metaspace_to_spatialdata
import spatialdata_plot # noqa: Not directly used but extends spatialdata
# Download dataset with optical background image
sdata = metaspace_to_spatialdata(
dataset_id="2022-08-05_17h28m56s",
fdr=0.1,
database=("BraChemDB", "2018-01"),
)
# Workaround: spatialdata-plot currently does not use points transformation
sdata.points["maldi_points"] = sdata.transform_element_to_coordinate_system(
sdata.points["maldi_points"], "global"
)
(
sdata.pl.render_images("optical_image")
.pl.render_points(
"maldi_points",
color=sdata.table.var.index[0],
alpha=1,
size=2,
cmap="viridis_r",
)
.pl.show(title=sdata.table.var.index[0], coordinate_systems="global")
)
Colocalization Analysis
A popular type of analysis for imaging Mass Spectrometry data or spatial metabolomics is colocalization analysis. In the ColocML publication, the authors evaluated data processing and colocalization metrics. Their best method (median filtering, quantile thresholding, and cosine similarity), can be executed with our package:
from metaspace_converter import metaspace_to_anndata, colocalization
# Download data
adata = metaspace_to_anndata(dataset_id="2023-11-14_21h58m39s", fdr=0.1)
# Perform median filtering and quantile thresholding
# The processed data is saved as a layer `adata.layers["coloc_ml_preprocessing"]`
# It has the same dimensions as `adata.X`
colocalization.coloc_ml_preprocessing(adata, layer="colocml_preprocessing")
# Compute the pairwise colocalization matrix between all ion images
# As an input, the processed data from `adata.layers["coloc_ml_preprocessing"]` is used
# The colocalization matrix is saved in `adata.varp["colocalization"]`
colocalization.colocalization(adata, layer="colocml_preprocessing")